publications
2025
-
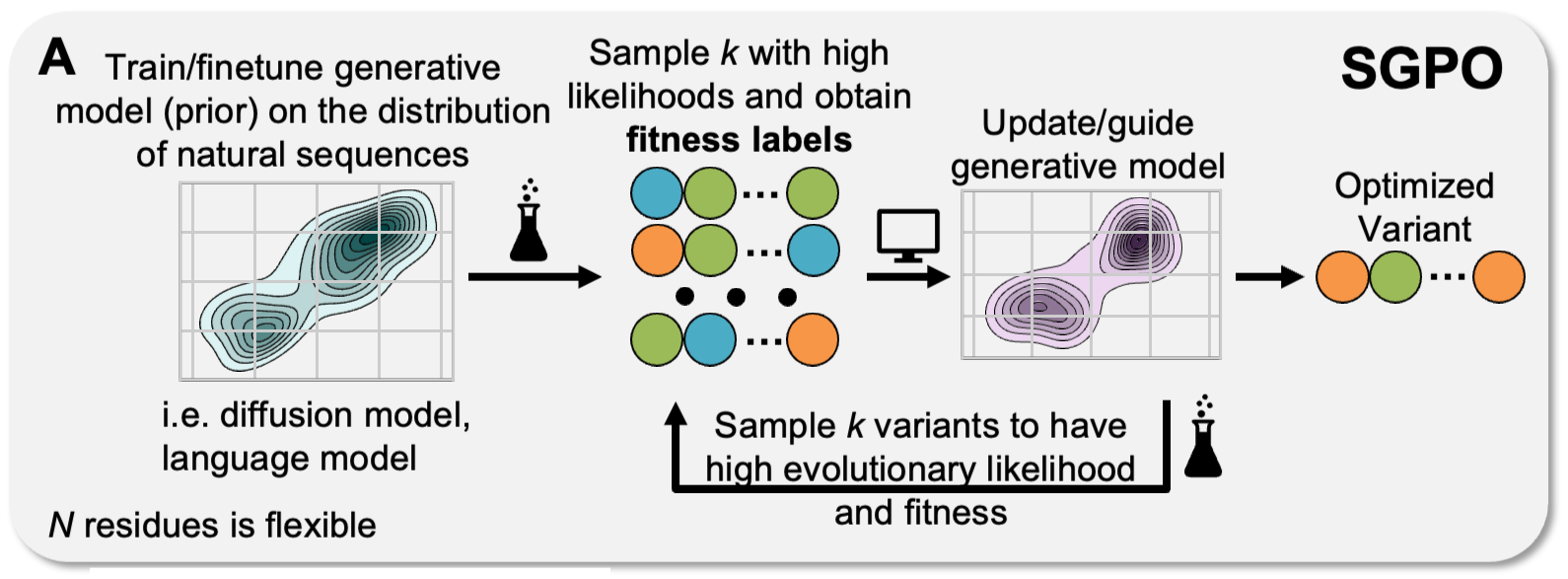 Steering Generative Models with Experimental Data for Protein Fitness OptimizationJason Yang, Wenda Chu, Daniel Khalil, and 4 more authorsNeurIPS, Dec 2025
Steering Generative Models with Experimental Data for Protein Fitness OptimizationJason Yang, Wenda Chu, Daniel Khalil, and 4 more authorsNeurIPS, Dec 2025Protein fitness optimization involves finding a protein sequence that maximizes desired quantitative properties in a combinatorially large design space of possible sequences. Recent developments in steering protein generative models (e.g diffusion models, language models) offer a promising approach. However, by and large, past studies have optimized surrogate rewards and/or utilized large amounts of labeled data for steering, making it unclear how well existing methods perform and compare to each other in real-world optimization campaigns where fitness is measured by low-throughput wet-lab assays. In this study, we explore fitness optimization using small amounts (hundreds) of labeled sequence-fitness pairs and comprehensively evaluate strategies such as classifier guidance and posterior sampling for guiding generation from different discrete diffusion models of protein sequences. We also demonstrate how guidance can be integrated into adaptive sequence selection akin to Thompson sampling in Bayesian optimization, showing that plug-and-play guidance strategies offer advantages compared to alternatives such as reinforcement learning with protein language models.
- Evaluation of Machine Learning-Assisted Directed Evolution Across Diverse Combinatorial LandscapesFrancesca-Zhoufan Li, Jason Yang, Kadina E Johnston, and 3 more authorsCell Systems, Sep 2025
Various machine learning-assisted directed evolution (MLDE) strategies have been shown to identify highfitness protein variants more efficiently than typical wet-lab directed evolution approaches. However, limited understanding of the factors influencing MLDE performance across diverse proteins has hindered optimal strategy selection for wet-lab campaigns. To address this, we systematically analyzed multiple MLDE strategies, including active learning and focused training using six distinct zero-shot predictors, across 16 diverse protein fitness landscapes. By quantifying landscape navigability with six attributes, we found that MLDE offers a greater advantage on landscapes which are more challenging for directed evolution, especially when focused training is combined with active learning. Despite varying levels of advantage across landscapes, focused training with zero-shot predictors leveraging distinct evolutionary, structural, and stability knowledge sources consistently outperforms random sampling for both binding interactions and enzyme activities. Our findings provide practical guidelines for selecting MLDE strategies for protein engineering.
- Sequence-Based Generative AI-Guided Design of Versatile Tryptophan SynthasesTheophile Lambert, Amin Tavakoli, Gautham Dharuman, and 5 more authorsbioRxiv, Aug 2025
Enzymes offer unparalleled selectivity and sustainability for chemical synthesis, yet their widespread industrial application is often hindered by the slow and uncertain process of discovering and optimizing suitable biocatalysts. While directed evolution remains the gold standard for enzyme optimization, its success hinges on the availability of a starting enzyme with measurable activity, a persistent bottleneck for many desired functions. Designing libraries likely to contain such functional starting points remains a major challenge. In this work, we use the GenSLM protein language model (PLM) along with a series of filters to generate novel sequences of the β-subunit of tryptophan synthase (TrpB) that express in Escherichia coli, are stable, and are catalytically active in the absence of a TrpA partner. Many generated TrpBs also demonstrated significant substrate promiscuity, accepting non-canonical substrates typically inaccessible to natural TrpBs. Remarkably, several outperformed both natural and laboratory-optimized TrpBs on native and non-canonical substrates. Comparative analysis of the most active and promiscuous generated TrpB and its closest natural homolog confirmed that this enhanced functional versatility does not stem from the natural enzyme, highlighting the creative potential of generative models. Our results demonstrate that the model can generate enzymes which not only preserve natural structure and function but also acquire non-natural properties, establishing PLMs as powerful tools for biocatalyst discovery and engineering, with the potential in some cases to bypass further optimization.
-
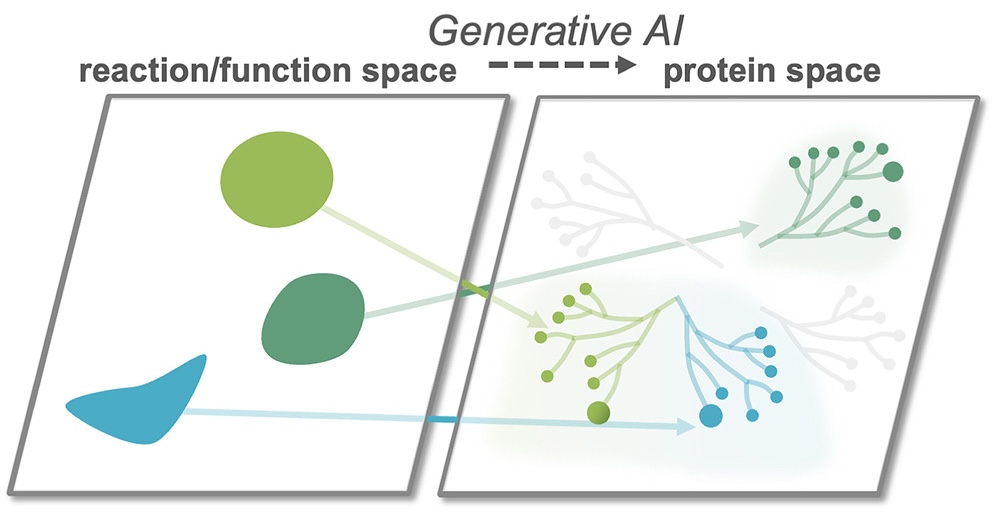 Illuminating the universe of enzyme catalysis in the era of artificial intelligenceJason Yang, Francesca-Zhoufan Li, Yueming Long, and 1 more authorCell Systems, Aug 2025
Illuminating the universe of enzyme catalysis in the era of artificial intelligenceJason Yang, Francesca-Zhoufan Li, Yueming Long, and 1 more authorCell Systems, Aug 2025Scientific research has revealed only a minuscule fraction of the enzymes that evolution has generated to power life’s essential chemical reactions—and an even tinier fraction of the vast universe of possible enzymes. Beyond the enzymes already annotated lie an astronomical number of biocatalysts that could enable sustainable chemical production, degrade toxic pollutants, and advance disease diagnosis and treatment. For the past few decades, directed evolution has been a powerful strategy for reshaping enzymes to access new chemical transformations: by harnessing nature’s existing diversity as a starting point and taking inspiration from nature’s most powerful design process, evolution, to modify enzymes incrementally. Recently, artificial intelligence (AI) methods have started revolutionizing how we understand and compose the language of life. In this perspective, we discuss a vision for AI-driven enzyme discovery to unveil a world of enzymes that transcends biological evolution and perhaps offers a route to genetically encoding almost any chemistry.
-
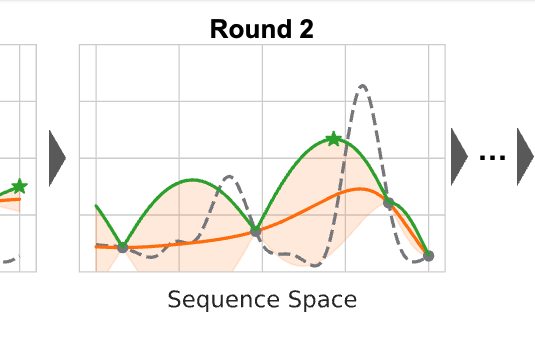 Active Learning-Assisted Directed EvolutionJason Yang, Ravi G Lal, James C Bowden, and 6 more authorsNature Communications, Jan 2025
Active Learning-Assisted Directed EvolutionJason Yang, Ravi G Lal, James C Bowden, and 6 more authorsNature Communications, Jan 2025Directed evolution (DE) is a powerful tool to optimize protein fitness for a specific application. However, DE can be inefficient when mutations exhibit non-additive, or epistatic, behavior. Here, we present Active Learning-assisted Directed Evolution (ALDE), an iterative machine learningassisted DE workflow that leverages uncertainty quantification to explore the search space of proteins more efficiently than current DE methods. We apply ALDE to an engineering landscape that is challenging for DE: optimization of five epistatic residues in the active site of an enzyme. In three rounds of wet-lab experimentation, we improve the yield of a desired product of a nonnative cyclopropanation reaction from 12% to 93%. We also perform computational simulations on existing protein sequence-fitness datasets to support our argument that ALDE can be more effective than DE. Overall, ALDE is a practical and broadly applicable strategy to unlock improved protein engineering outcomes.
2024
-
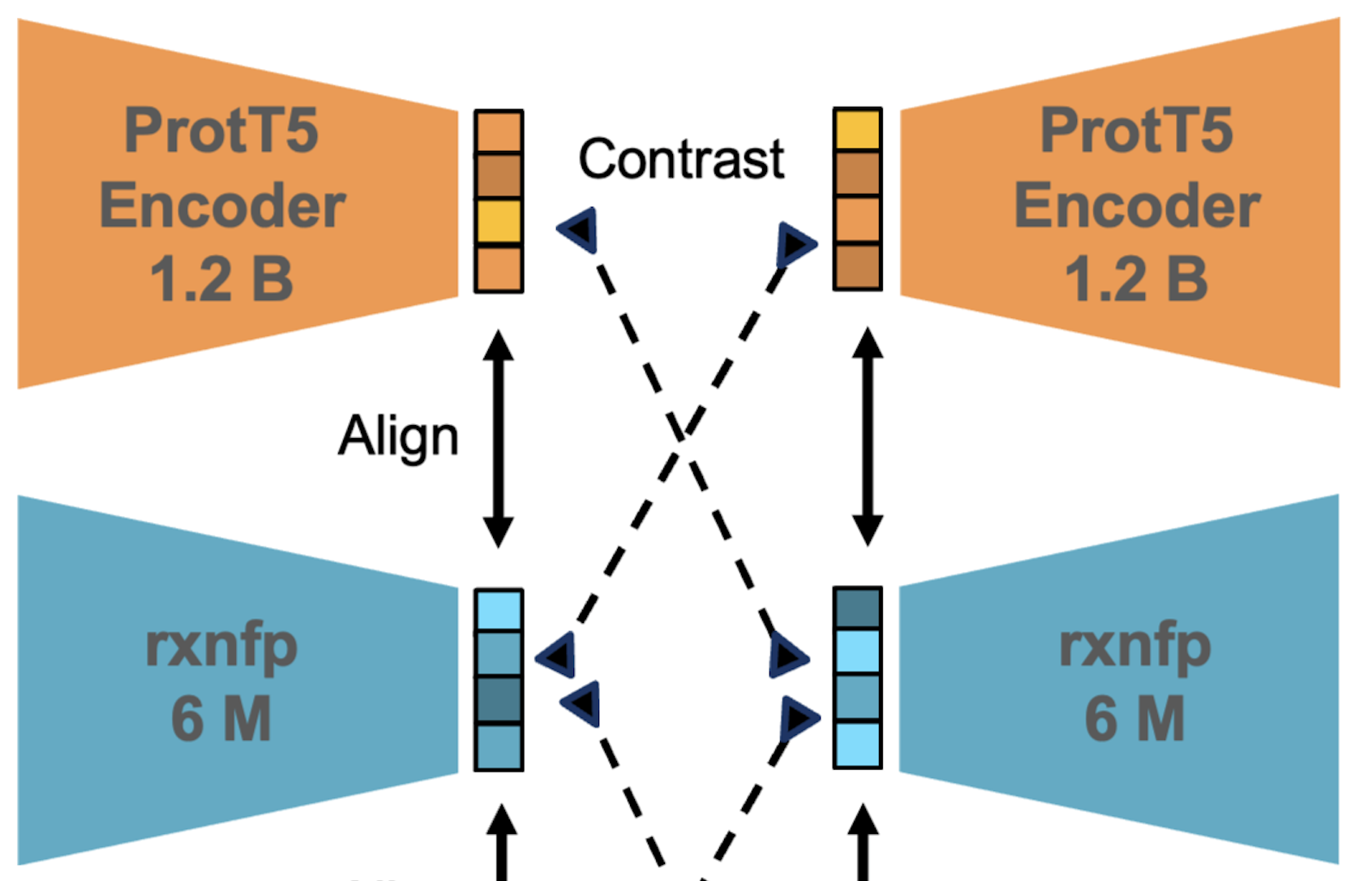 CARE: a Benchmark Suite for the Classification and Retrieval of EnzymesJason Yang, Ariane Mora, Shengchao Liu, and 4 more authorsNeurIPS, Dec 2024
CARE: a Benchmark Suite for the Classification and Retrieval of EnzymesJason Yang, Ariane Mora, Shengchao Liu, and 4 more authorsNeurIPS, Dec 2024Enzymes are important proteins that catalyze chemical reactions. In recent years, machine learning methods have emerged to predict enzyme function from sequence; however, there are no standardized benchmarks to evaluate these methods. We introduce CARE, a benchmark and dataset suite for the Classification And Retrieval of Enzymes (CARE). CARE centers on two tasks: (1) classification of a protein sequence by its enzyme commission (EC) number and (2) retrieval of an EC number given a chemical reaction. For each task, we design train-test splits to evaluate different kinds of out-of-distribution generalization that are relevant to real use cases. For the classification task, we provide baselines for state-of-the-art methods. Because the retrieval task has not been previously formalized, we propose a method called Contrastive Reaction-EnzymE Pretraining (CREEP) as one of the first baselines for this task and compare it to the recent method, CLIPZyme. CARE is available at https://github.com/jsunn-y/CARE/.
-
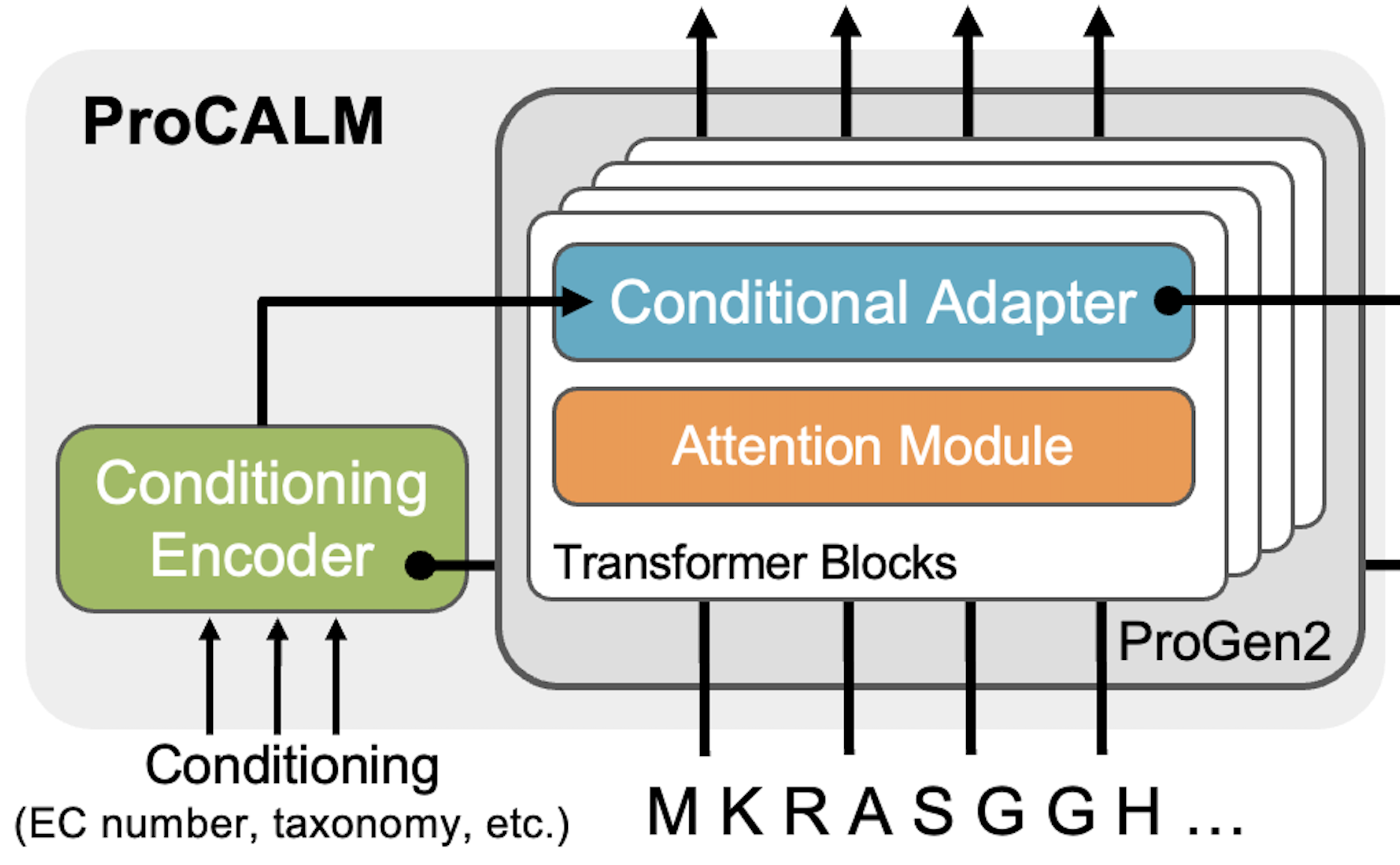 Function-Guided Conditional Generation Using Protein Language Models with AdaptersJason Yang, Aadyot Bhatnagar, Jeffrey A. Ruffolo, and 1 more authorMLSB Workshop, Dec 2024
Function-Guided Conditional Generation Using Protein Language Models with AdaptersJason Yang, Aadyot Bhatnagar, Jeffrey A. Ruffolo, and 1 more authorMLSB Workshop, Dec 2024The conditional generation of proteins with desired functions is a key goal for generative models. Existing methods based on prompting of protein language models (PLMs) can generate proteins conditioned on a target functionality, such as a desired enzyme family. However, these methods are limited to simple, tokenized conditioning and have not been shown to generalize to unseen functions. In this study, we propose ProCALM (Protein Conditionally Adapted Language Model), an approach for the conditional generation of proteins using adapters to PLMs. While previous methods have used adapters for structure-conditioned generation from PLMs, our implementation of ProCALM involves finetuning ProGen2 to condition generation based on versatile representations of protein function-e.g. enzyme family, taxonomy, or natural language descriptions. ProCALM matches or exceeds the performance of existing methods at conditional sequence generation from target functions. Impressively, it can also generalize to rare and unseen functions. Overall, ProCALM is a flexible and computationally efficient approach, and we expect that it can be extended to a wide range of generative language models.
-
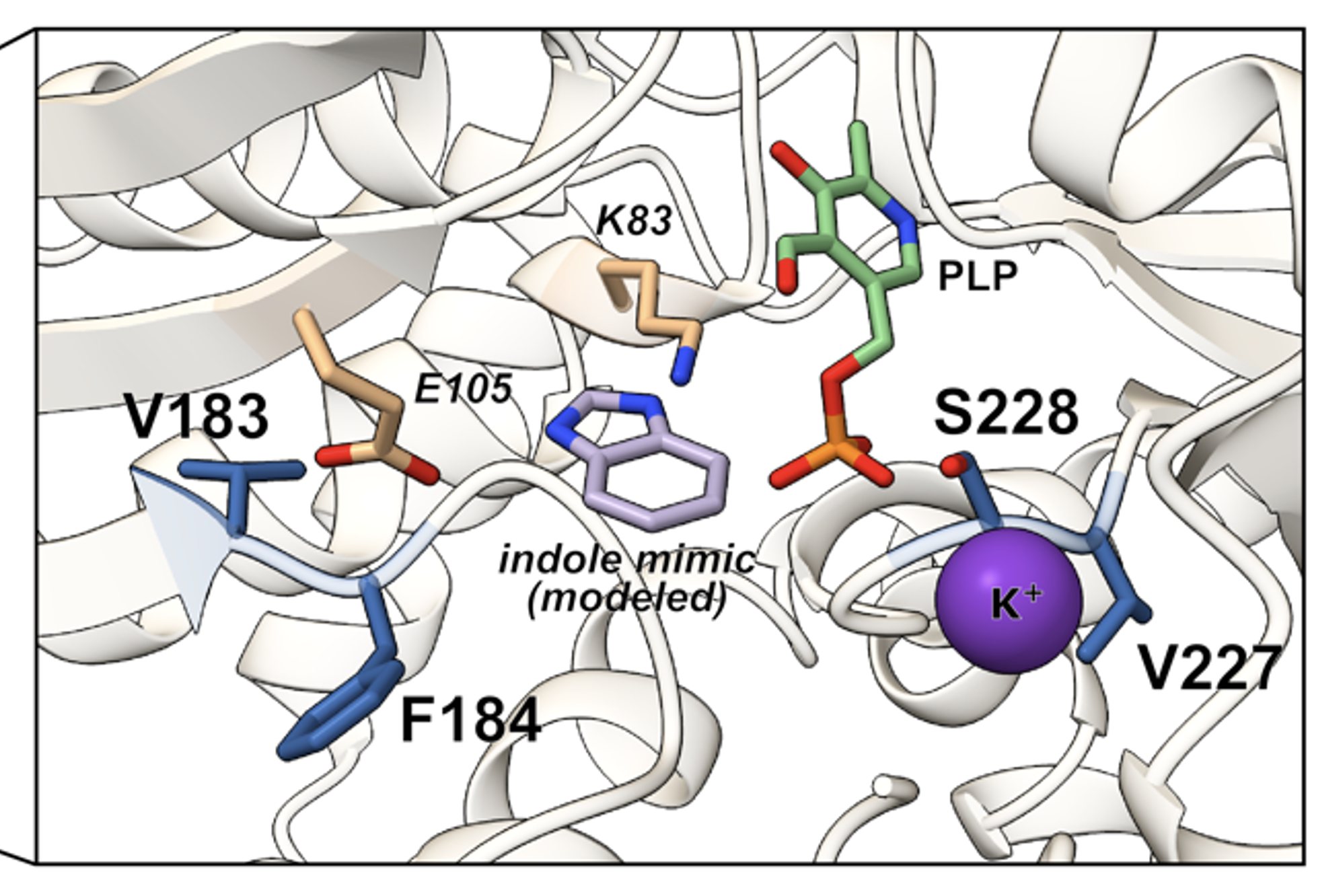 A combinatorially complete epistatic fitness landscape in an enzyme active siteKadina E Johnston, Patrick J Almhjell, Ella J Watkins-Dulaney, and 4 more authorsPNAS, Jul 2024
A combinatorially complete epistatic fitness landscape in an enzyme active siteKadina E Johnston, Patrick J Almhjell, Ella J Watkins-Dulaney, and 4 more authorsPNAS, Jul 2024Protein engineering often targets binding pockets or active sites which are enriched in epistasis—non-additive interactions between amino acid substitutions—and where the combined effects of multiple single substitutions are difficult to predict. Few existing sequence-fitness datasets capture epistasis at large scale, especially for enzyme catalysis, limiting the development and assessment of model-guided enzyme engineering approaches. We present here a combinatorially complete, 160,000-variant fitness landscape across four residues in the active site of an enzyme. Assaying the native reaction of a thermostable β-subunit of tryptophan synthase (TrpB) in a non-native environment yielded a landscape characterized by significant epistasis and many local optima. These effects prevent simulated directed evolution approaches from efficiently reaching the global optimum. There is nonetheless wide variability in the effectiveness of different directed evolution approaches, which together provide experimental benchmarks for computational and machine learning workflows. The most-fit TrpB variants contain a substitution that is nearly absent in natural TrpB sequences—a result that conservationbased predictions would not capture. Thus, although fitness prediction using evolutionary data can enrich in more-active variants, these approaches struggle to identify and differentiate among the most-active variants, even for this near-native function. Overall, this work presents a new, large-scale testing ground for model-guided enzyme engineering and suggests that efficient navigation of epistatic fitness landscapes can be improved by advances in both machine learning and physical modeling.
-
 Opportunities and Challenges for Machine Learning-Assisted Enzyme EngineeringJason Yang, Francesca-Zhoufan Li, and Frances H. ArnoldACS Central Science, Feb 2024
Opportunities and Challenges for Machine Learning-Assisted Enzyme EngineeringJason Yang, Francesca-Zhoufan Li, and Frances H. ArnoldACS Central Science, Feb 2024Enzymes can be engineered at the level of their amino acid sequences to optimize key properties such as expression, stability, substrate range, and catalytic efficiency�or even to unlock new catalytic activities not found in nature. Because the search space of possible proteins is vast, enzyme engineering usually involves discovering an enzyme starting point that has some level of the desired activity followed by directed evolution to improve its “fitness” for a desired application. Recently, machine learning (ML) has emerged as a powerful tool to complement this empirical process. ML models can contribute to (1) starting point discovery by functional annotation of known protein sequences or generating novel protein sequences with desired functions and (2) navigating protein fitness landscapes for fitness optimization by learning mappings between protein sequences and their associated fitness values. In this Outlook, we explain how ML complements enzyme engineering and discuss its future potential to unlock improved engineering outcomes.
2023
-
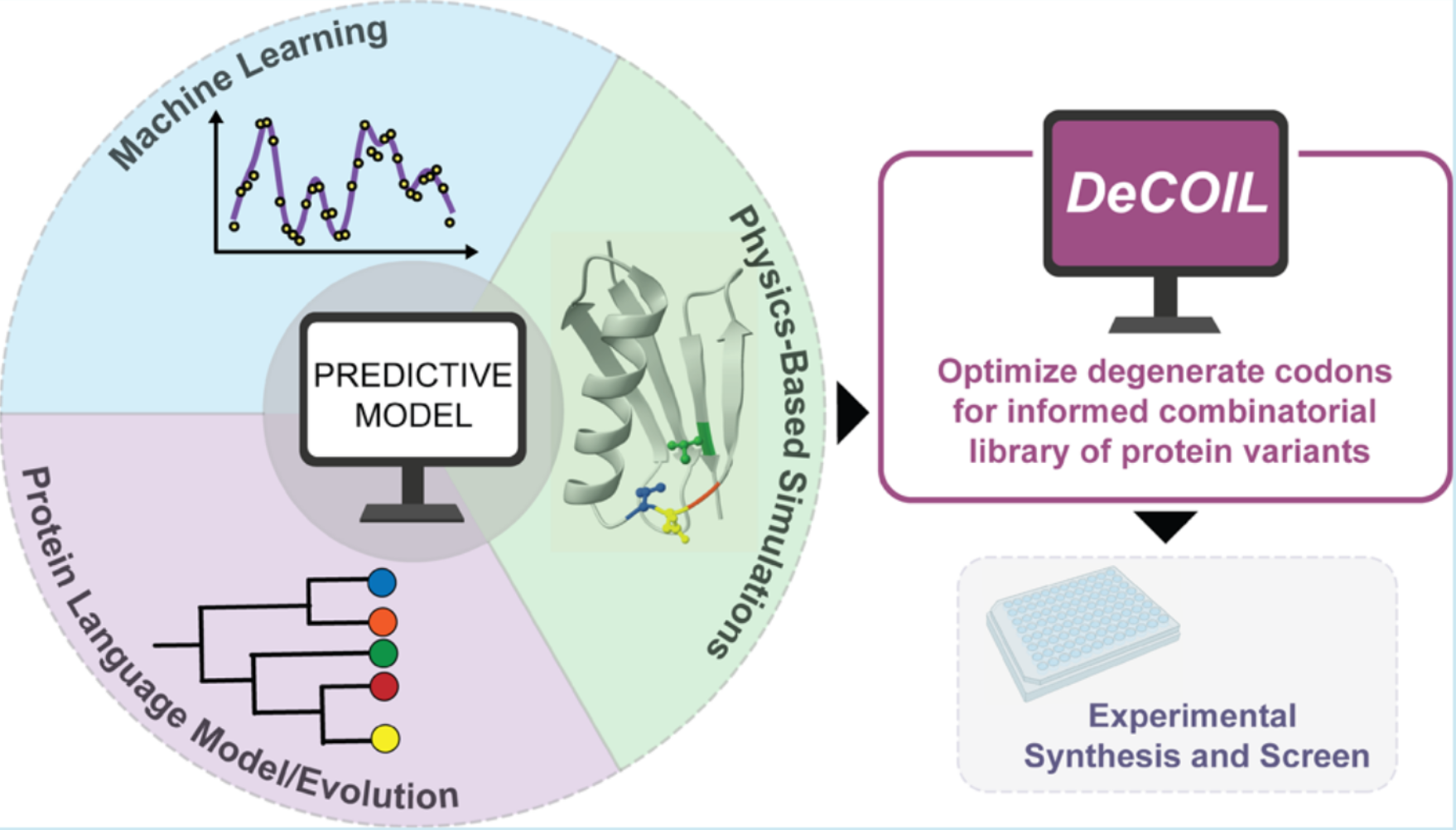 DeCOIL: Optimization of Degenerate Codon Libraries for Machine Learning-Assisted Protein EngineeringJason Yang, Julie Ducharme, Kadina E. Johnston, and 3 more authorsACS Synthetic Biology, Jul 2023
DeCOIL: Optimization of Degenerate Codon Libraries for Machine Learning-Assisted Protein EngineeringJason Yang, Julie Ducharme, Kadina E. Johnston, and 3 more authorsACS Synthetic Biology, Jul 2023With advances in machine learning (ML)-assisted protein engineering, models based on data, biophysics, and natural evolution are being used to propose informed libraries of protein variants to explore. Synthesizing these libraries for experimental screens is a major bottleneck, as the cost of obtaining large numbers of exact gene sequences is often prohibitive. Degenerate codon (DC) libraries are a cost-effective alternative for generating combinatorial mutagenesis libraries where mutations are targeted to a handful of amino acid sites. However, existing computational methods to optimize DC libraries to include desired protein variants are not well suited to design libraries for ML-assisted protein engineering. To address these drawbacks, we present DEgenerate Codon Optimization for Informed Libraries (DeCOIL), a generalized method that directly optimizes DC libraries to be useful for protein engineering: to sample protein variants that are likely to have both high fitness and high diversity in the sequence search space. Using computational simulations and wet-lab experiments, we demonstrate that DeCOIL is effective across two specific case studies, with the potential to be applied to many other use cases. DeCOIL offers several advantages over existing methods, as it is direct, easy to use, generalizable, and scalable. With accompanying software (https://github.com/jsunn-y/DeCOIL), DeCOIL can be readily implemented to generate desired informed libraries.
2022
-
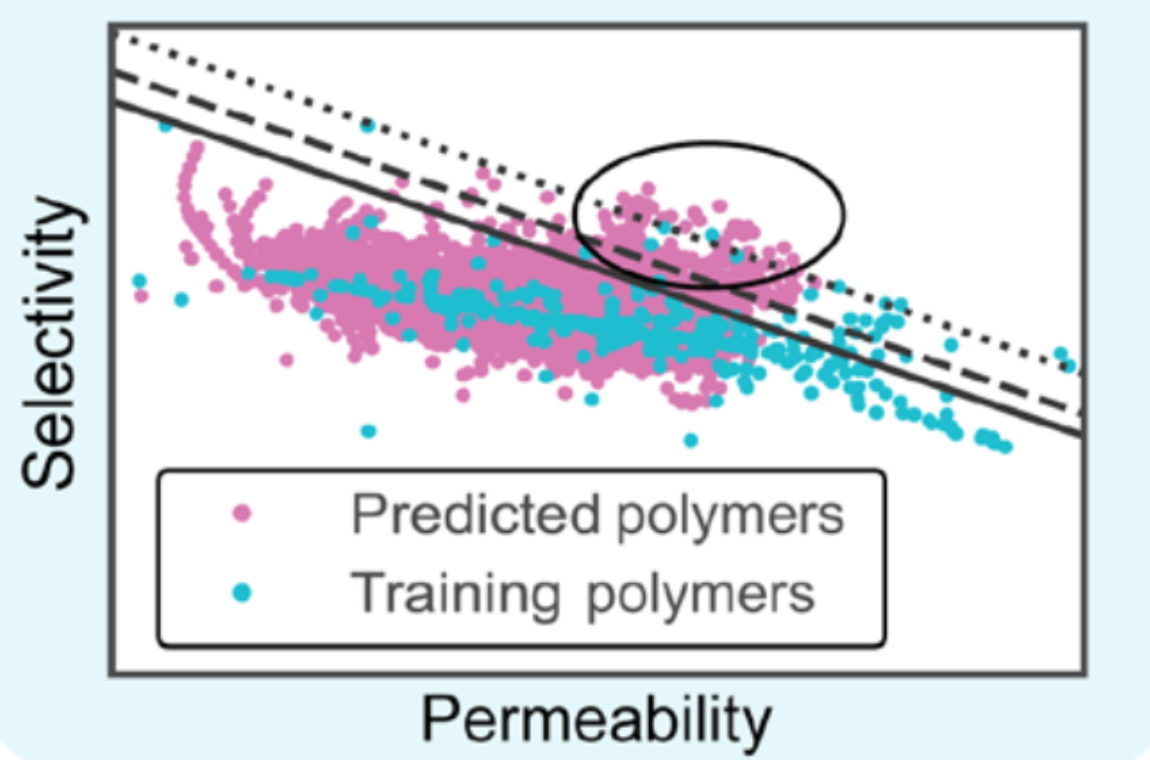 Machine learning enables interpretable discovery of innovative polymers for gas separation membranesJason Yang, Lei Tao, Jinlong He, and 2 more authorsScience Advances, Jul 2022
Machine learning enables interpretable discovery of innovative polymers for gas separation membranesJason Yang, Lei Tao, Jinlong He, and 2 more authorsScience Advances, Jul 2022Polymer membranes perform innumerable separations with far-reaching environmental implications. Despite decades of research, design of new membrane materials remains a largely Edisonian process. To address this shortcoming, we demonstrate a generalizable, accurate machine learning (ML) implementation for the discovery of innovative polymers with ideal performance. Specifically, multitask ML models are trained on experimental data to link polymer chemistry to gas permeabilities of He, H 2 , O 2 , N 2 , CO 2 , and CH 4 . We interpret the ML models and extract valuable insights into the contributions of different chemical moieties to permeability and selectivity. We then screen over 9 million hypothetical polymers and identify thousands that lie well above current performance upper bounds, including hundreds of never-before-seen ultrapermeable polymer membranes with O 2 and CO 2 permeability greater than 10 4 and 10 5 Barrers, respectively. High-fidelity molecular dynamics simulations confirm the ML-predicted gas permeabilities of the promising candidates, which suggests that many can be translated to reality. , Machine learning generates chemical insights for the design of polymeric gas separation membranes with exceptional performance.
- Molecular insights into the structure-property relationships of 3D printed polyamide reverse-osmosis membrane for desalinationJinlong He, Jason Yang, Jeffrey R. McCutcheon, and 1 more authorJournal of Membrane Science, Sep 2022
3D-printing is an emerging method for manufacturing polyamide (PA) reserve osmosis (RO) membranes for water treatment and desalination, which can precisely control membrane structural properties, such as thickness, roughness, and resolution. However, the synthesis-structure (i.e., degree of cross-linking (DC), m-phenylenediamine/trimesoyl chloride (MPD/TMC) ratio, and membrane thickness) to property (permeability and water-salt selectivity) relationships for these membranes has not been well understood. At the same time, a microscopic understanding of the physical mechanism of water and salt transport is needed to guide the design of high-performance 3D-printed membranes and improve the printing efficiency. Thus, the atomic-scale transport features and energetics of water and salt ions are studied at high pressure for the 3D-printed PA RO membranes with the different DCs and MPD/TMC ratios through non-equilibrium molecular dynamics (NEMD) simulations. Factoring in membrane structure properties, rejection ratio of salt ions and pressure-dependent water flux, 3D-printed PA membranes having an MPD/TMC ratio of 3.0:2.0 and a DC between 80%∼90% attains ideal performance: high water flux, high rejection of salt ions, and excellent structural integrity. Mechanistically, water permeability for highly cross-linked PA RO membranes depends on the temporary on-and-off channels that allow water molecules to jump from one cavity to another at high pressure. In addition, higher pressures cause rapid compaction of PA membranes’ free volume and membrane thickness. Membrane failure at high pressure is determined by the DC and MPD/TMC ratios-dependent compressive yield strength. In short, these findings provide physical insights for optimizing existing PA membranes and designing next-generation desalination membranes at the molecular level.
- Designing polymeric membranes with coordination chemistry for high-precision ion separationsRyan M. DuChanois, Mohammad Heiranian, Jason Yang, and 5 more authorsScience Advances, Mar 2022
State-of-the-art polymeric membranes are unable to perform the high-precision ion separations needed for technologies essential to a circular economy and clean energy future. Coordinative interactions are a mechanism to increase sorption of a target species into a membrane, but the effects of these interactions on membrane permeability and selectivity are poorly understood. We use a multilayered polymer membrane to assess how ion-membrane binding energies affect membrane permeability of similarly sized cations: Cu 2+ , Ni 2+ , Zn 2+ , Co 2+ , and Mg 2+ . We report that metals with higher binding energy to iminodiacetate groups of the polymer more selectively permeate through the membrane in multisalt solutions than single-salt solutions. In contrast, weaker binding species are precluded from diffusing into the polymer membrane, which leads to passage proportional to binding energy and independent of membrane thickness. Our findings demonstrate that selectivity of polymeric membranes can markedly increase by tailoring ion-membrane binding energy and minimizing membrane thickness. , Tailoring interactions between a target ion and polymer can lead to highly precise ion separations for ultrathin membranes.
2021
-
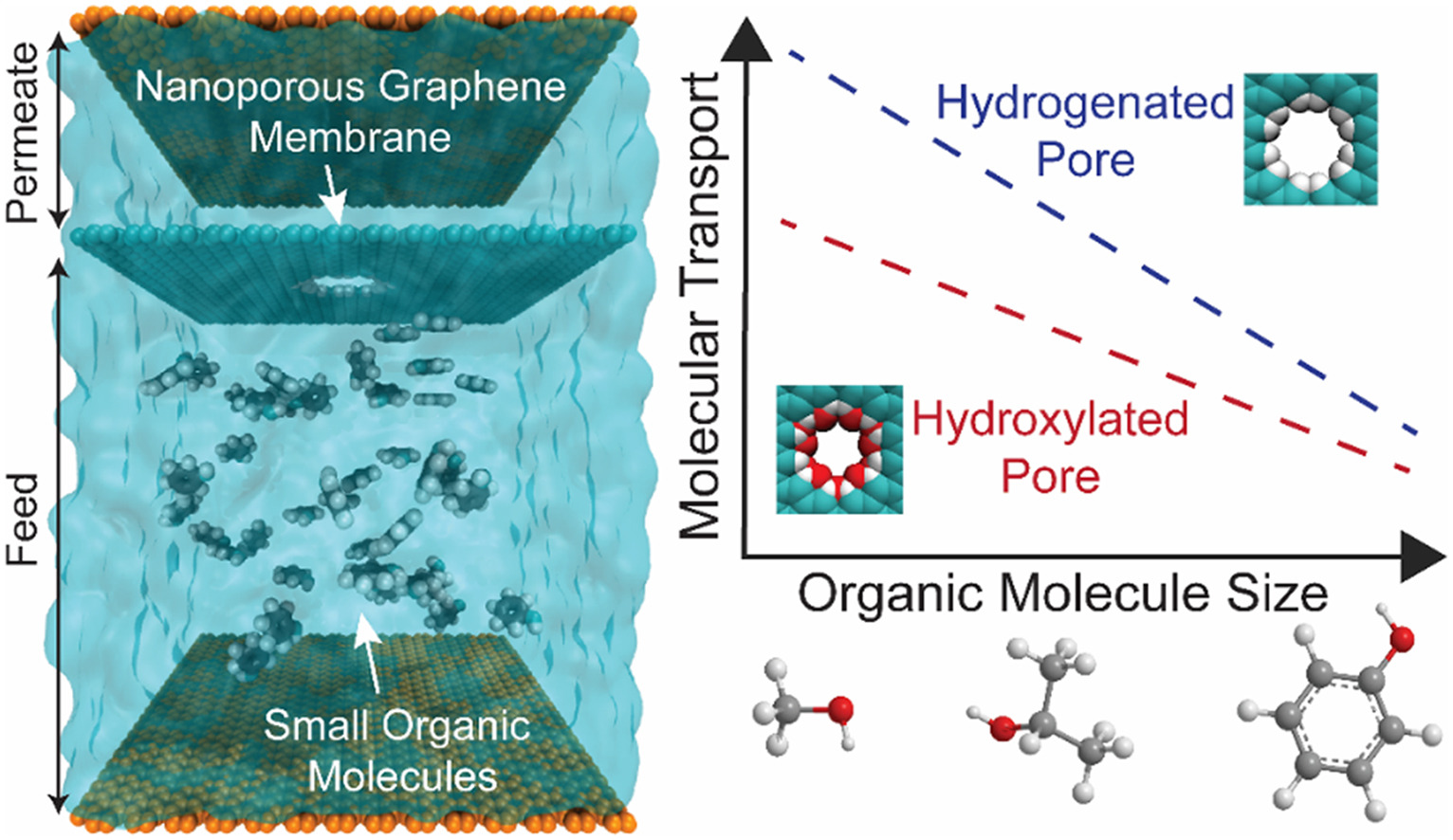 Efficient separation of small organic contaminants in water using functionalized nanoporous graphene membranes: Insights from molecular dynamics simulationsJason Yang, Zhiqiang Shen, Jinlong He, and 1 more authorJournal of Membrane Science, Jul 2021
Efficient separation of small organic contaminants in water using functionalized nanoporous graphene membranes: Insights from molecular dynamics simulationsJason Yang, Zhiqiang Shen, Jinlong He, and 1 more authorJournal of Membrane Science, Jul 2021Small organic molecules, and specifically micropollutants, pose known hazards to human health, but their removal is particularly challenging in water treatment. Our work demonstrates promising single-layer nanoporous graphene (NPG) membranes with high water permeability and varying degrees of selectivity against common organic contaminants, using molecular dynamics (MD) simulations. Seven target organic molecules are considered—including methanol, urea, ethanol, 2-propanol, n-nitrosodimethylamine (NDMA), pyrrole, and phenol—to understand the molecular parameters that govern organic removal. We systematically study molecular transport dynamics and energetics through membranes having varying sizes of pores with hydrophobic (hydrogenated) and hydrophilic (hydroxylated) functionalizations. We find that NPG membranes with smaller, hydroxylated pores offer higher water/organic permselectivity compared to those with larger, hydrogenated pores, as they impede the transport of organics while facilitating the transport of water. Molecular size is the primary organic parameter that controls transport. Larger organic molecules have a greater affinity for the membrane and pore groups (interfacial-affinity sieving), but they are more hindered from entering the pore (pore-size sieving). There is a net decrease in transport with increased molecular size, which suggests that pore-size sieving is the governing mechanism for most of the molecular separations. We further find that traditional measures of organic hydrophobicity are not strong predictors of transport, as they correlate poorly with interfacial affinity, and the contribution of dehydration to the energy barrier of permeation is limited, though non-negligible. Finally, our simulations show that organic-pore interactions are not pressure independent: as flow rate is decreased, the effects of interfacial-affinity sieving become more pronounced. In short, this study establishes a comprehensive understanding of membrane and molecular parameters to guide the design of effective NPG membranes for organic contaminant removal, particularly in desalination and wastewater reuse.
2019
-
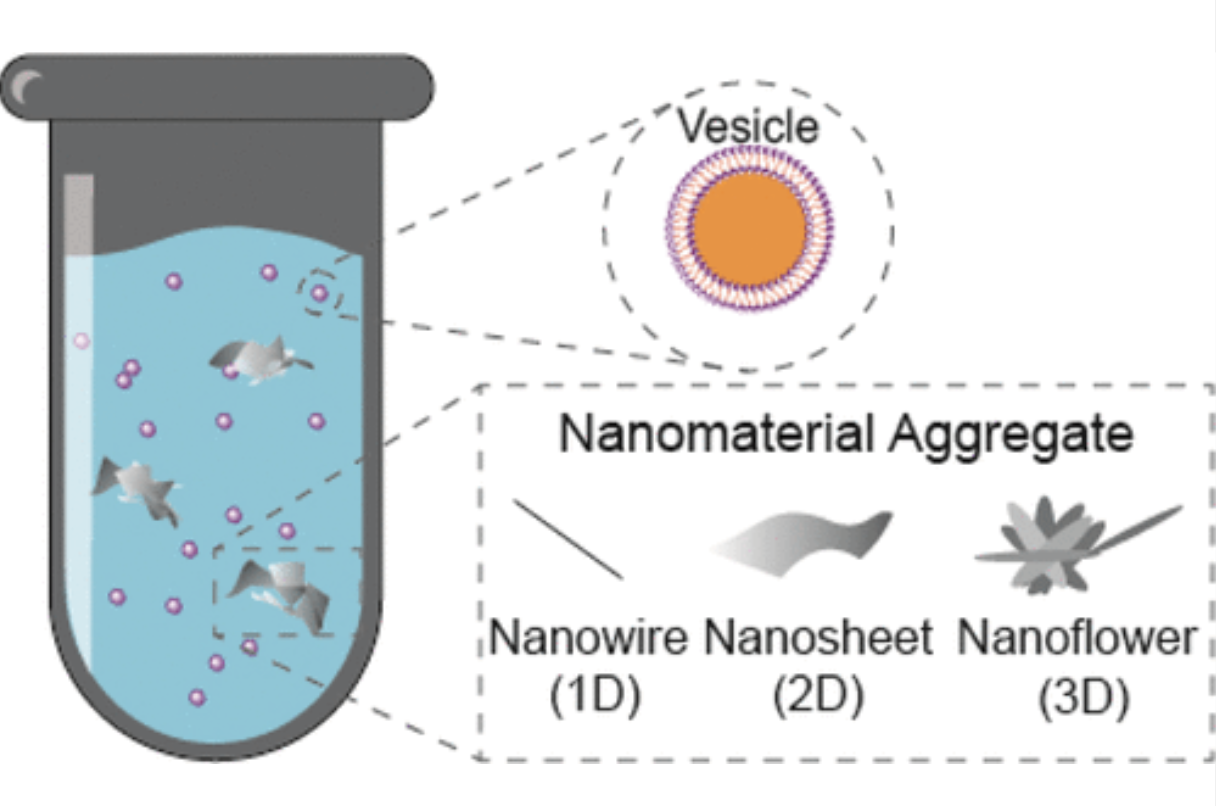 Shape-Dependent Interactions of Manganese Oxide Nanomaterials with Lipid Bilayer VesiclesInes Zucker, Sara M. Hashmi, Jason Yang, and 3 more authorsLangmuir, Oct 2019
Shape-Dependent Interactions of Manganese Oxide Nanomaterials with Lipid Bilayer VesiclesInes Zucker, Sara M. Hashmi, Jason Yang, and 3 more authorsLangmuir, Oct 2019Interactions of transition-metal-oxide nanomaterials with biological membranes have important environmental implications and applications in ecotoxicity and life-cycle assessment analysis. In this study, we quantitatively assess the impact of MnO2 nanomaterial morphology—one-dimensional (1D) nanowires, 2D nanosheets, and 3D nanoflowers—on their interaction with phospholipid vesicles as a model for biological membranes. Confocal microscopy suggests visual evidence for the interaction of undisrupted vesicles with dispersed MnO2 nanomaterials of different morphologies, and it further supports the observation that minimal dye leakage of the vesicle inner solution was detected during the interaction with MnO2 nanomaterials during the dye leakage assay. Upon titration of vesicles to dispersions of MnO2 nanowires, nanosheets, and nanoflowers, each roughly 10 times larger than the vesicles, dynamic light scattering reveals two diffusive time scales associated with aggregates in the mixture. While the longer time scale corresponds to the dispersed MnO2 control population, the appearance of a shorter timescale with vesicle addition indicates interaction between the dispersed metal oxide nanomaterials and the vesicles. The interaction is shape-dependent, being more pronounced for MnO2 nanowires than for nanosheets and nanoflowers. Furthermore, the shorter diffusive time scale is intermediate between the vesicle and nanomaterial controls, which may suggest a degree of metal oxide aggregate breakup. Vesicle adsorption isotherms and zeta potential measurements during titration corroborate vesicle attachment on the nanomaterials. Our results suggest that the dispersed nanomaterial shape plays an important role in mediating nondestructive vesicle–nanomaterial interactions and that lipid vesicles act as efficient surfactants for MnO2 nanomaterials.